Covariate-adjusted heatmaps for visualizing biological data via correlation decomposition
Han-Ming Wu1,Yin-Jing Tien2, Meng-Ru Ho3, 4, 5, Hai-Gwo Hwu6, Wen-chang Lin5, Mi-Hua Tao5, and Chun-Houh Chen2,*
1 Department of Statistics, National Taipei University, New Taipei
City, 23741,
Taiwan, R.O.C.,
2Institute of Statistical Science, Academia Sinica, Taipei 11529,
Taiwan, R.O.C.,
3Institute of Biomedical Informatics, National Yang-Ming University,
Taipei 112, Taiwan, R.O.C.,
4Bioinformatics Program, Taiwan International Graduate Program,
Academia Sinica, Taipei 115, Taiwan, R.O.C.,
5Institute of Biomedical Sciences, Academia Sinica, Taipei 115,
Taiwan, R.O.C.,
6Department of Psychiatry, National Taiwan University Hospital and
College of Medicine, Taipei 100, Taiwan, R.O.C.
and Department of Psychology, College of Public Health, Neurobiology and
Cognitive Science Center, Taipei 100, Taiwan, R.O.C.
*to whom correspondence should be addressed.
The psychosis disorder data
¡@
1. WABA and z-score significance map
(1.1) Import the psychosis disorder data
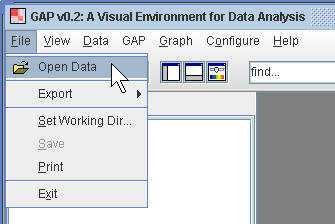
¡@
(1.2) In the Open Data Dialog Window, specify the column name and the covariate.
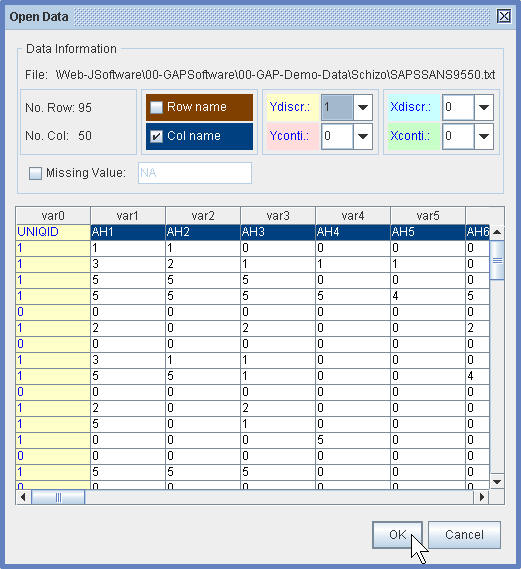
¡@
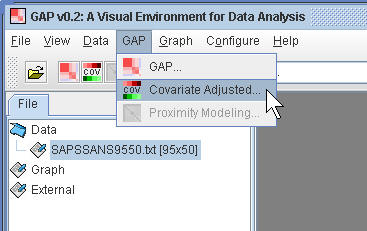
(1.3) Click the menu for Covariate Adjusted.
(1.4) In the Decomposition of Correlation Matrices Dialog Window, specify the covariate to be adjusted.
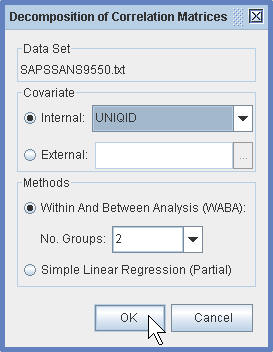
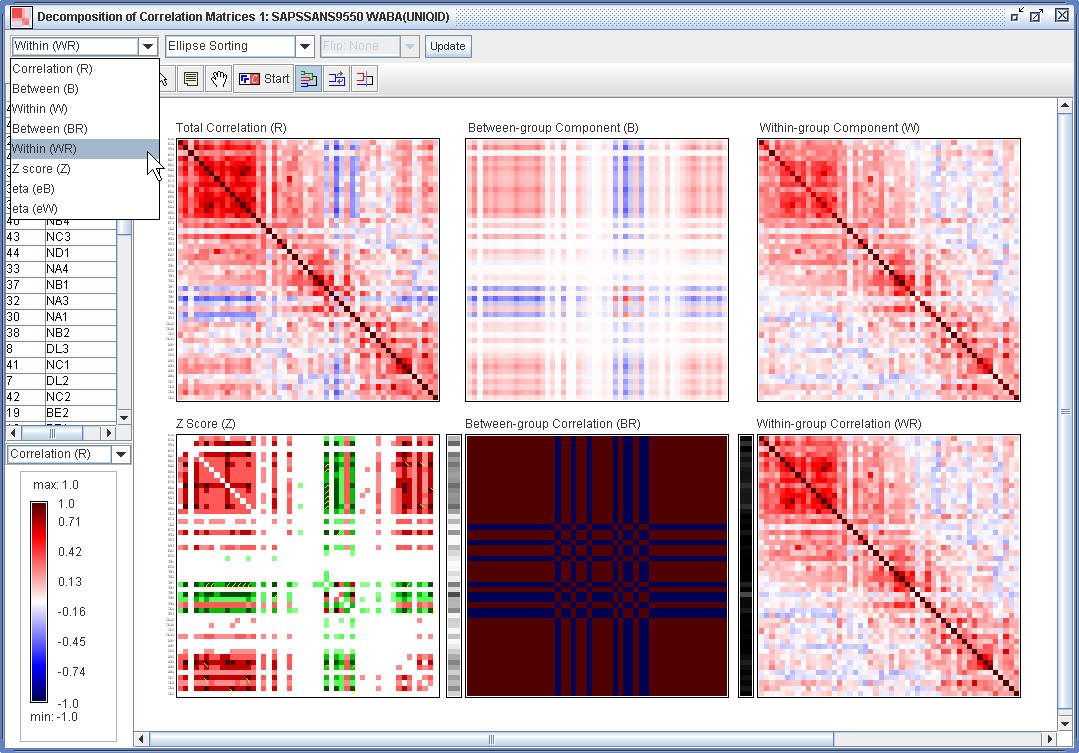
(1.5) The the WABA Window, one can reorder the rows and columns of the specified matrix using HCT or R2E.
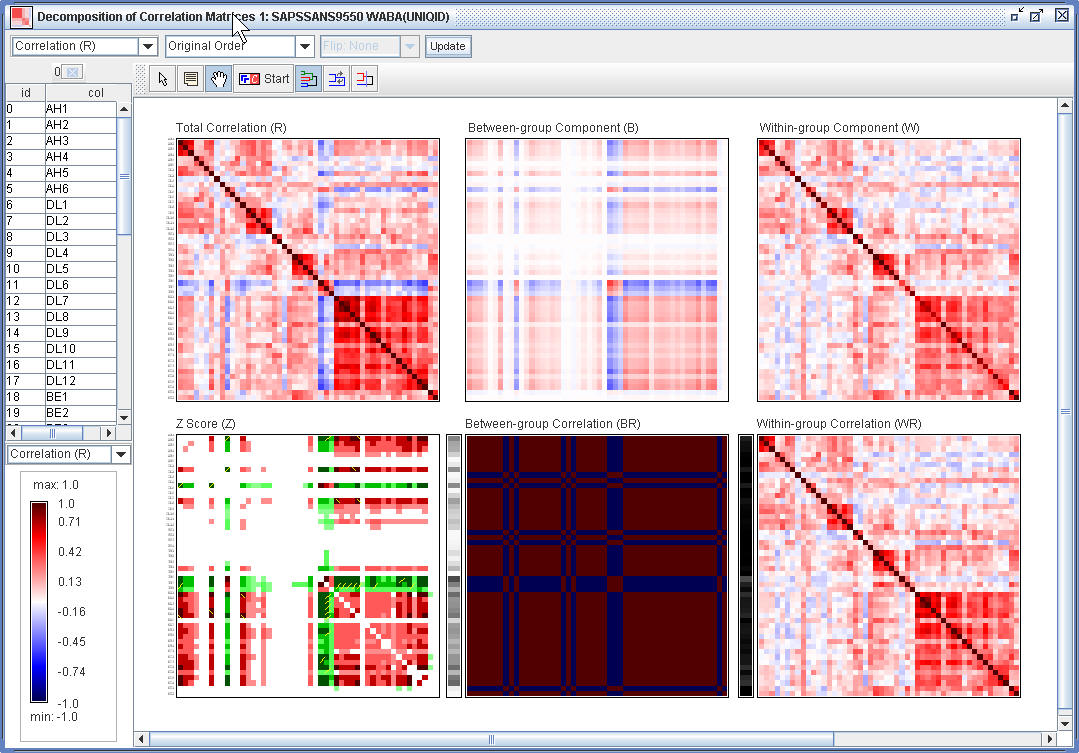
(1.6) Sort the total correlation map using R2E.
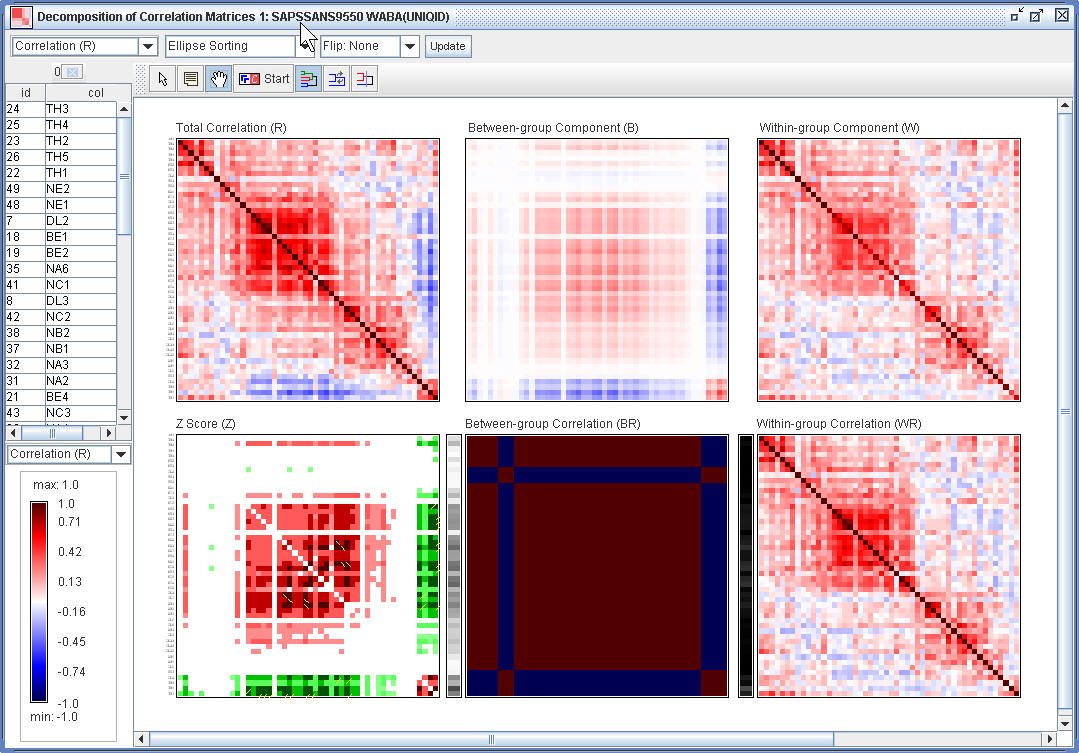
¡@
(1.7) Sort the adjusted correlation map (WR) using R2E.
¡@
¡@
2. Covariate-adjusted MV
(2.1) Click the menu for GAP analysis.
![]()
¡@
(2.2) In the Generalized Association Plots Dialog Window, specify the data type, the proximity measures and the covariate adjustment option.
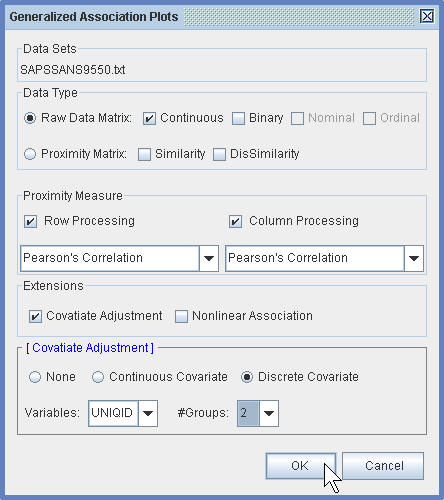
¡@
(2.3) In the Generalized Association Plots Window, use the control panel to zoom and coloring. Select the Residual Data option.
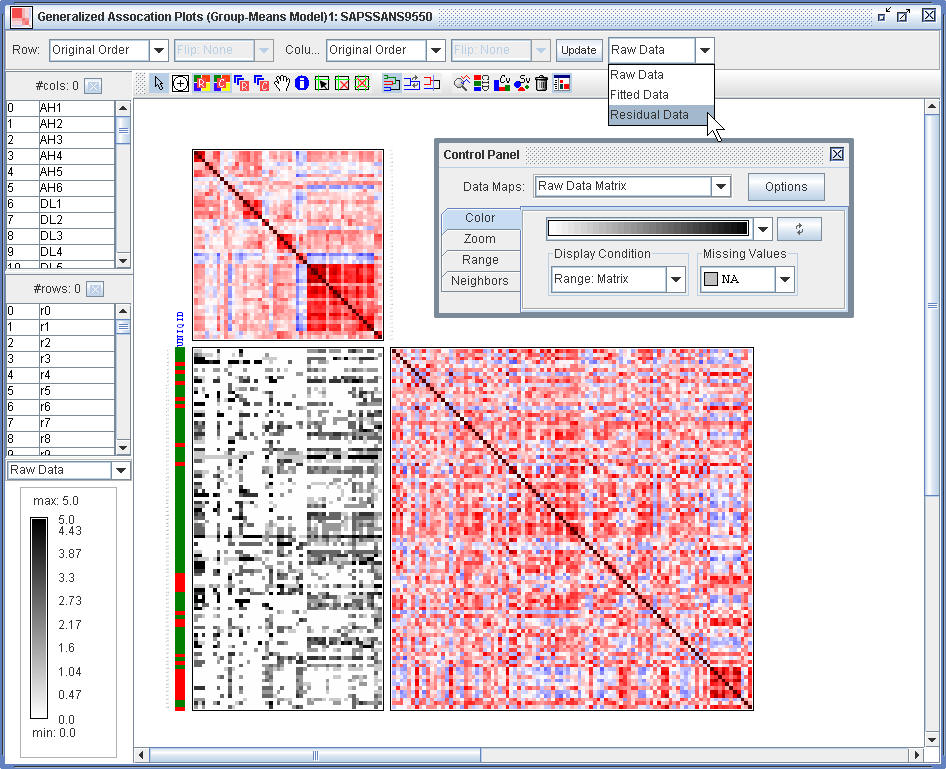
¡@
(2.4) One can do the GAP analysis for the residual data.
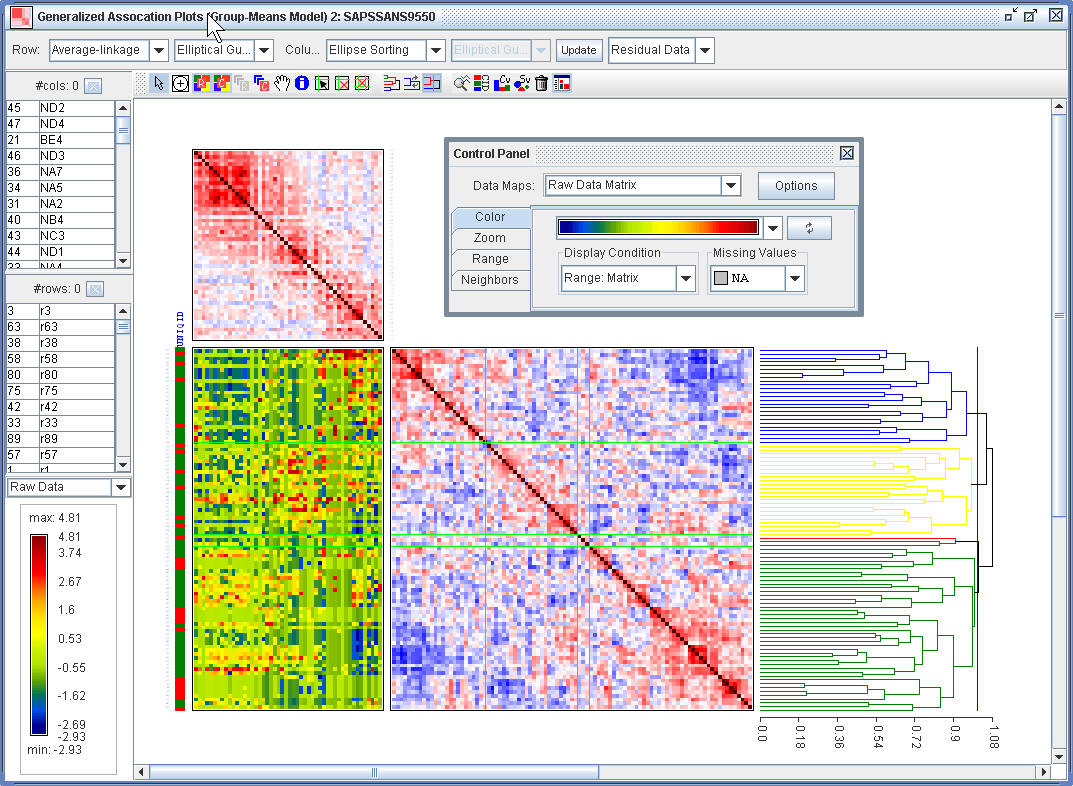
¡@
(2.5) One can also do the GAP analysis for the fitted data.
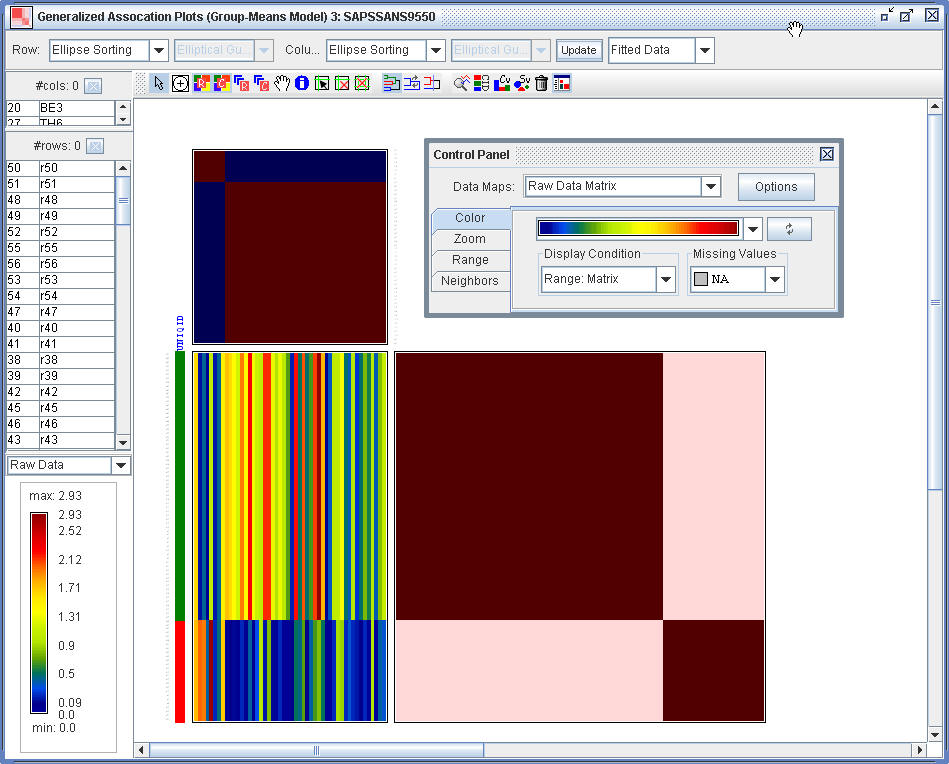
Last Update: 2018/03/15, hmwu@gm.ntpu.edu.tw